Introduction
The SET binding protein 1 (SETBP1) gene is an oncogene located on chromosome 18q12.3, which is related to DNA replication and gene transcription regulation. Variants in SETBP1 are involved in germline and somatic mutations, leading to extremely different pathologic consequences. Germline mutations in SETBP1 are associated with Schinzel-Giedion syndrome (SGS) and SETBP1 Haploinsufficiency Disorder. In contrast, somatic mutations in SETBP1 are linked with myeloid malignancies and clonal hematopoiesis. The vast majority of reported germline SETBP1 mutations were located in the SKI homologous region (codons 868-871). However, germline SETBP1 mutations outside the hotspot region were rarely reported and the potential carcinogenicity remained unclear.
Methods
Patients from 2018-2022 at the Hennan Cancer Hospital were identified. Bone marrow samples at diagnosis of 285 sporadic patients with myeloid malignancies were sequenced using next generation sequencing (NGS). The candidate SETBP1 mutations were validated by Sanger sequencing from bone marrow and exfoliated buccal epithelial cell samples. Whole exome sequencing (WES) was performed on patient 5 and her parents from the family with SETBP1 mutations.
Results
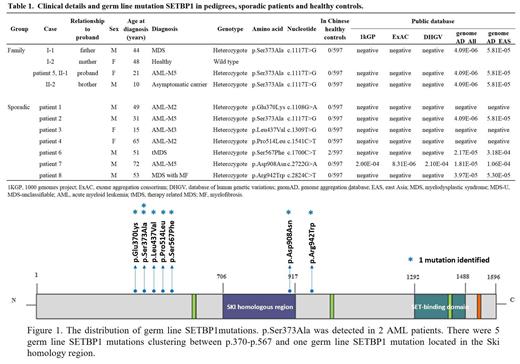
In our study, 8 (2.8%) patients (6 AML and 2 MDS) were identified with germ line SETBP1 mutations (Table 1, Figure 1). Importantly, patient 2 and patient 5 carried the same germ line SETBP1 p.Ser373Ala mutation. Meanwhile, 4 patients had somatic SETBP1 mutations at hotspots (p.Asp868Asn in 1 patient and p.Gly870Ser in 3 patients). None of these germ line SETBP1 mutations was detected in 597 healthy Chinese individuals except p.Ser567Phe (2 out of 597) (Table 1). Minimum allele frequencies (MAF) of the germ line SETBP1 mutations were negligible in public databases (Table 1). A significant difference ( p<0.05) was observed in the minimum allele frequency (MAF) of germ line SETBP1 mutations between sporadic MDS/AML patients and healthy controls, which suggested that germ line SETBP1 mutations should be closely related to MDS/AML.
Furthermore, we investigated the family history of the patients with germ line SETBP1 mutation. The father of patient 5 was diagnosed with MDS and carried germ line SETBP1 p.Ser373Ala mutation. Known germ line mutations or somatic mutations associated with myeloid malignancies were not detected in the family of patient 5 by WES. Patient 2, who also carried the germ line SETBP1 p.Ser373Ala mutation, had no family history of myeloid disorders. The elder sister (carrier) of patient 2 had persistent abnormal CBC while the little sister (carrier) had normal CBC. Somatic SETBP1 mutations conveyed a poor prognosis in myeloid malignancies. And in our data, patient 5 and patient 2 both appeared to be induction failure and early relapse. Then patient 2 underwent HLA-full-matched allo-HSCT in the second remission and the doner was his little sister. Unfortunately, after allo-HSCT the CR only maintained 2 months. None of the other 6 sporadic MDS/AML patients with germ line SETBP1 mutation had a family history of hematologic disorders. Many family members were detected to carry germ line SETBP1 mutations but they did not show the characteristics of myeloid disorders other than abnormal percentages of neutrophils and lymphocytes.
Conclusion
In our study, we identified novel germ line SETBP1 mutations in 8 sporadic MDS/AML patients, and a family history of MDS/AML was noted in one. Germ line SETBP1 mutations are not infrequent among non-familial MDS/AML patients. Identification of these cases is imperative especially in patients who are candidates for HSCT from a family donor, since the association between germ line SETBP1 mutations and recipient outcomes was uncertain.
Disclosures
Liu:Rigel: Speakers Bureau; Beigene: Speakers Bureau.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal